Hi Everyone,
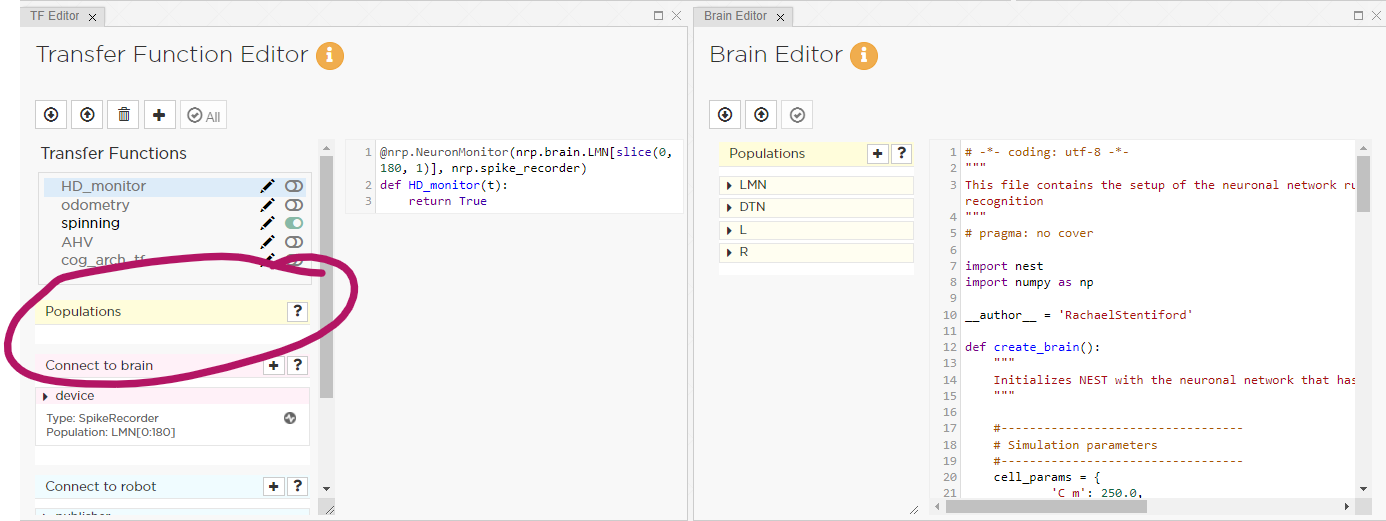
I am having a problem consistently when launching an experiment on the NRP. When i launch my experiment the transfer function editor cannot see any of the populations defined in the brain model editior.
The experiment was originally cloned from the WhiskEye template and the bibi file altered to accept a NEST model rather than a PYNN model.
It looks like the brain model is not recognised initially. If i try to add a new population i get an error: ‘No brain is currently loaded, cannot add Populations’.
If i change something in the brain editor and apply those changes, i can then add new populations.
Only once i have applied a change and added a new population is the transfer function editor able to see the populations.
This is frustrating for me, but also makes this experiment unusable as a demonstrator. Does anyone have any suggestions as to how to fix this issue?
Cheers
Rachael